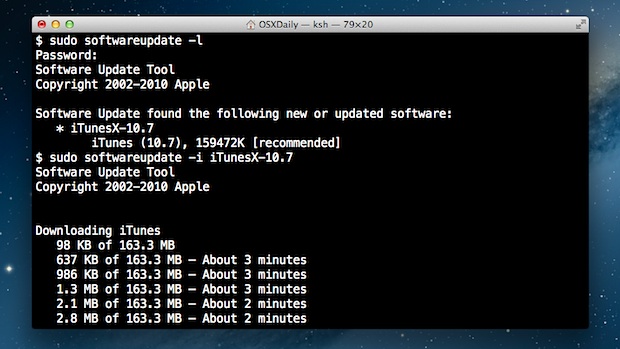
Xcode-select: error: command line tools are already installed, use “Software Update” to install updates. This means you'll need to go to the developer website instead. Installing the CLT. When the.dmg has finished downloaded, double click the file to open it. This will open a little window that looks like this. Install Command Line Tools. Open your terminal to begin the process of installing command-line tools on macOS Catalina. With your terminal opened, type the command below into the prompt and press the return key to execute it. After executing the command below a pop-up window should open with the option to install. The default SVN version which is installed along with Xcode command line tools is 1.7.x. If you're fine with this version, than that should be enough. I want to select my SVN version and for that I'm using Homebrew.
Os X Command Line Tools
- Global Nav Open Menu Global Nav Close Menu; Apple; Shopping Bag +.
- If you're also facing The 'xcode-select' command requires the command line developer tools error then watch this video because in this video I'm going to sho.
BLAST+ executables
Do you have difficulties running high volume BLAST searches? Do you have proprietary sequence data to search and cannot use the NCBI BLAST web site? Do you have access to your own server? Do you have your own research pipeline? Have security or IP concerns about sending searches outside of your organization? If you answered yes to any of these questions, read on!
The NCBI provides a suite of command-line tools to run BLAST called BLAST+. This allows users to perform BLAST searches on their own server without size, volume and database restrictions. BLAST+ can be used with a command line so it can be integrated directly into your workflow. Case sv250 operator manual.
What are the next steps?
Download and install BLAST+. Installers and source code are available from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/. Notes app word count. Download the databases you need,(see database section below), or create your own. Start searching.
_1605536569.png)
For more details, please see the BLAST+ user manual, the BLAST Help manual, the BLAST releases notes, and the article in BMC Bioinformatics (PubMed link). Cla vocals stereo link to. See our versioning policy.
Command Line Tools Download For Mac
The BLAST+ suite is the currently supported package. The older C toolkit executables are no longer supported. See our versioning policy.
We are always listening and welcome your feedback at BLAST Support Center.
Magic-BLAST
Mac Command Line Tools Install
Magic-BLAST is a tool for mapping large next-generation RNA or DNA sequencing runs against a whole genome or transcriptome. Read more about Magic-BLAST at https://ncbi.github.io/magicblast.
Installers and source code are available from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/magicblast/LATEST.
IgBLAST
IgBLAST facilitates the analysis of immunoglobulin and T cell receptor variable domain sequences. For details please see the documentation at https://ncbi.github.io/igblast/ and the article in Nucleic Acids Research (https://www.ncbi.nlm.nih.gov/pubmed/23671333).
Installers and source code are available from ftp://ftp.ncbi.nih.gov/blast/executables/igblast/release/LATEST.
SRPRISM
SRPRISM is a short read alignment tool that works with genomic sequences and handles alternative loci. For more information, see ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/srprism/README. A LINUX executable is available under ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/srprism

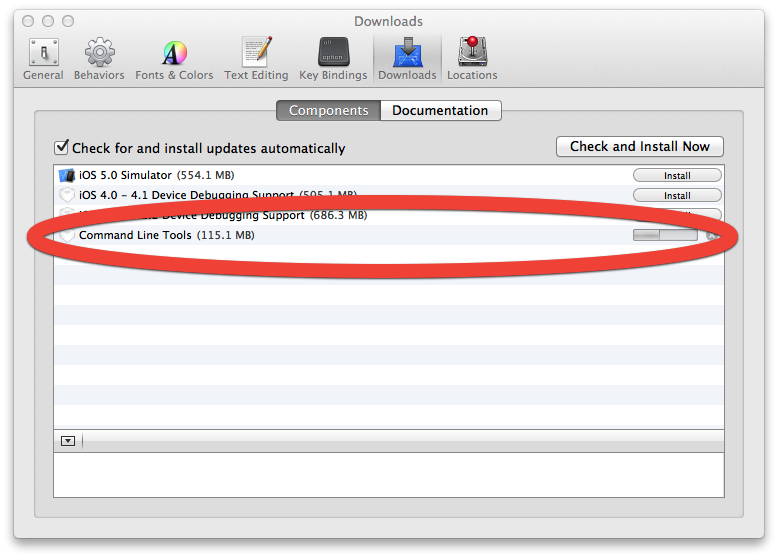
Download Mac Os Command Line Tools

Databases
BLAST databases are updated daily and may be downloaded via FTP fromftp://ftp.ncbi.nlm.nih.gov/blast/db/.Database sets may be retrieved automatically with update_blastdb.pl, which is part of the BLAST+ suite.Please refer to the BLAST database documentation for more details.
Downloading Command Line Tools (macos High Sierra Version 10.13) For Xcode
The NCBI makes searchable collection of position-specific scoring matrices that can be used for sensitive protein and translated nucleotide searches. These are known as the Conserved Domain Database and can be searched with the RPSBLAST and RPSTBLASTN executables distributed with the BLAST+ package. The most comprehensive collection of CDD's is available at ftp://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/little_endian/Cdd_NCBI_LE.tar.gz. Subsets of the comprehensive collection are available at ftp://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/little_endian/. Documentation on these CDD collections is available at ftp://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/README.



